Welcome to Viroverse
Viroverse is a platform for the collection, storage, retrieval, and analysis of biological samples and experimental data for laboratory workflows. Developed in-house for sixteen years, it serves as the principal data store for HIV sequencing experiments conducted in the Mullins Lab. Viroverse currently houses a repository listing for hundreds of thousands of specimens and hundreds of thousands of viral nucleotide sequences, together with comprehensive metadata about their creation including PCR protocols, gel images, subject clinical data, and more.
Want to try Viroverse for your own work? You can obtain the source code from GitHub. If you want further support, please contact the Mullins Lab to discuss the possibilities.
Complete molecular virology workflow
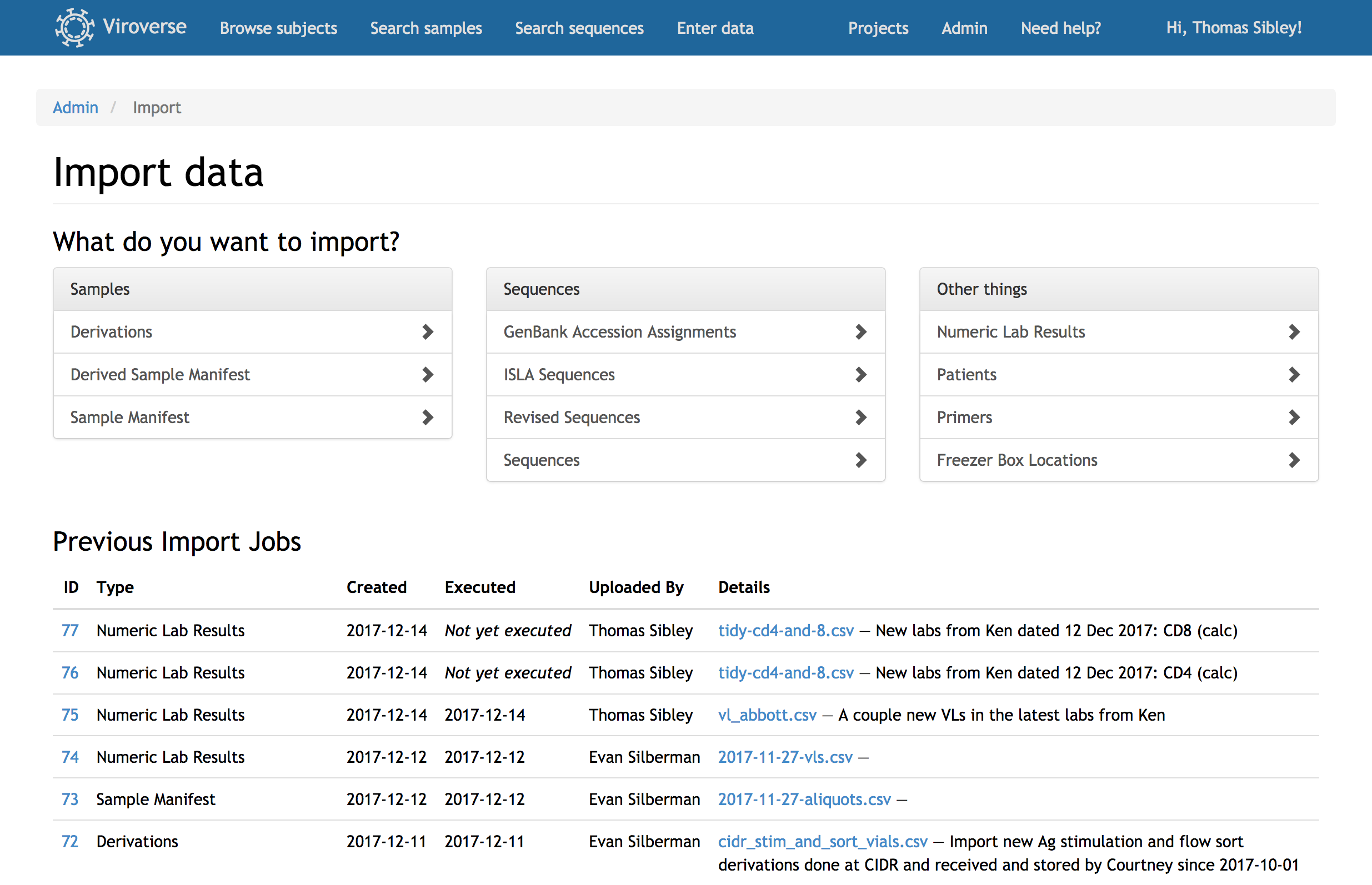
Track the provenance of viral sequences starting from the original tissue sample. Each workflow step—nucleic acid extraction, reverse transcription, PCR, and sequencing—is recorded, including protocol details like primers, enzymes, and output concentrations.
Fast and accessible search
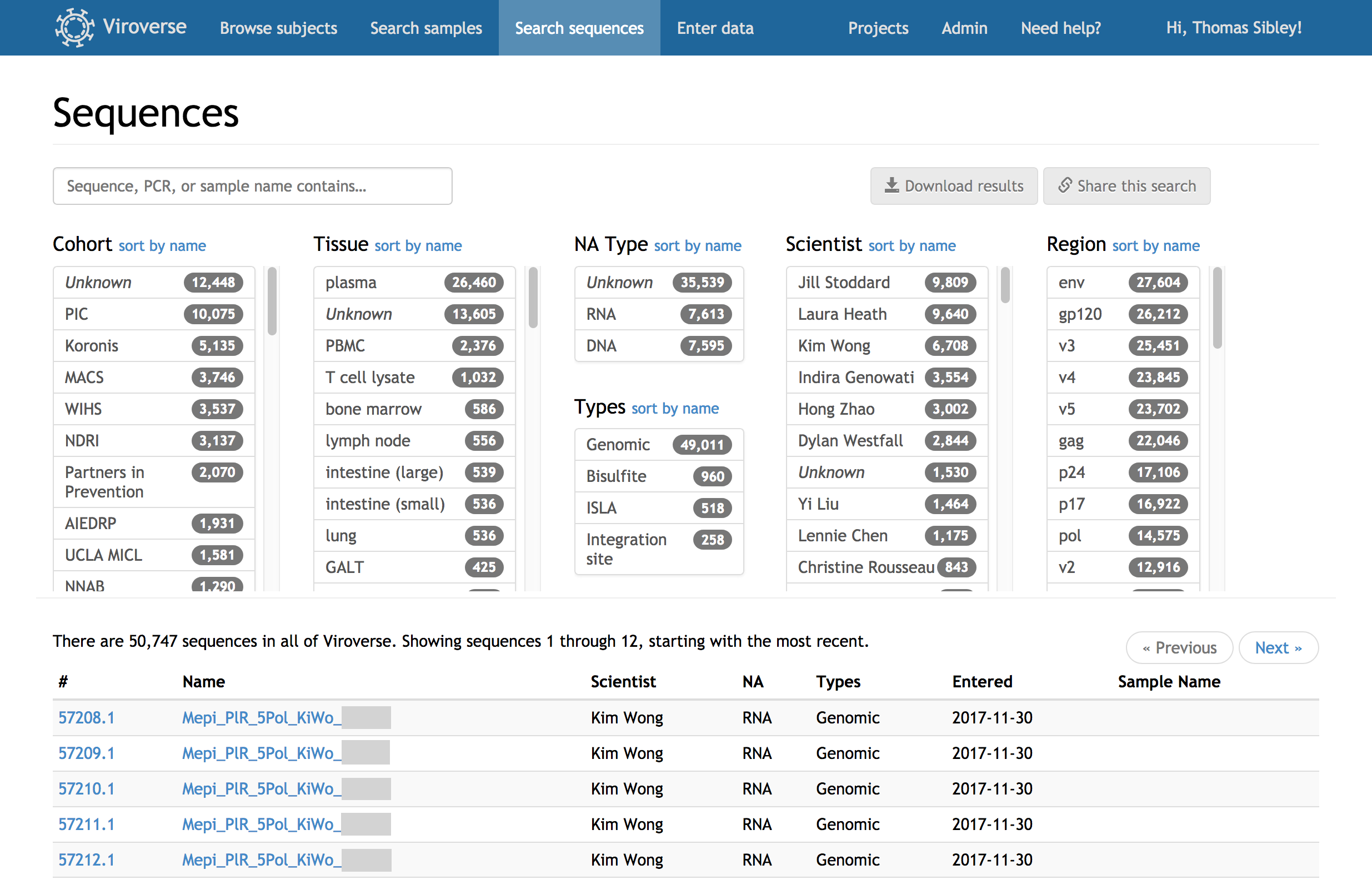
Pull specific sequences and samples from a collection of thousands in seconds. Viroverse’s search allows filtering by tissue, project, scientist, amplicon, and more. Integration with Viroblast lets you match sequence contents against your whole collection to screen for contamination.
Longitudinal cohort tracking
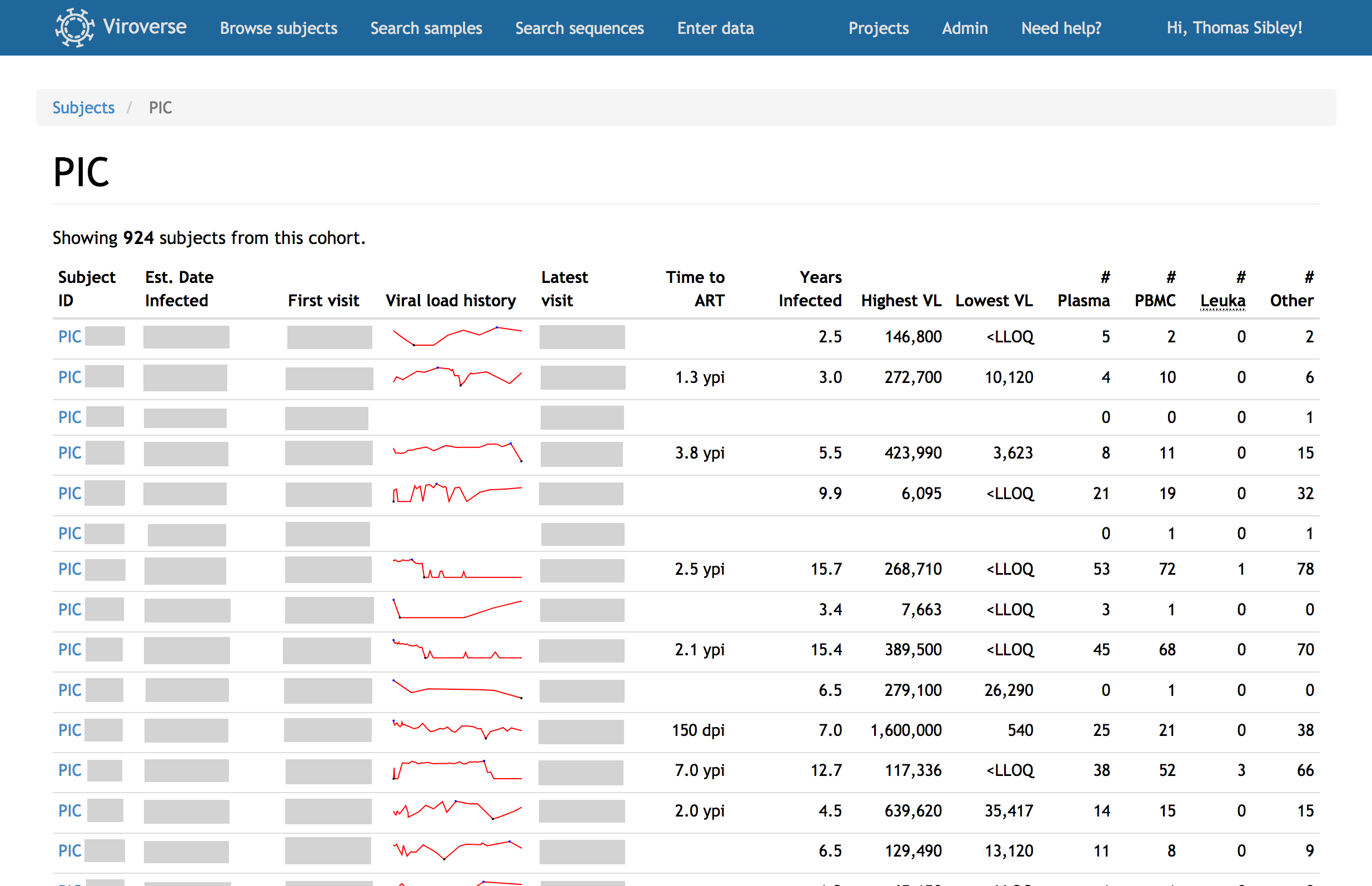
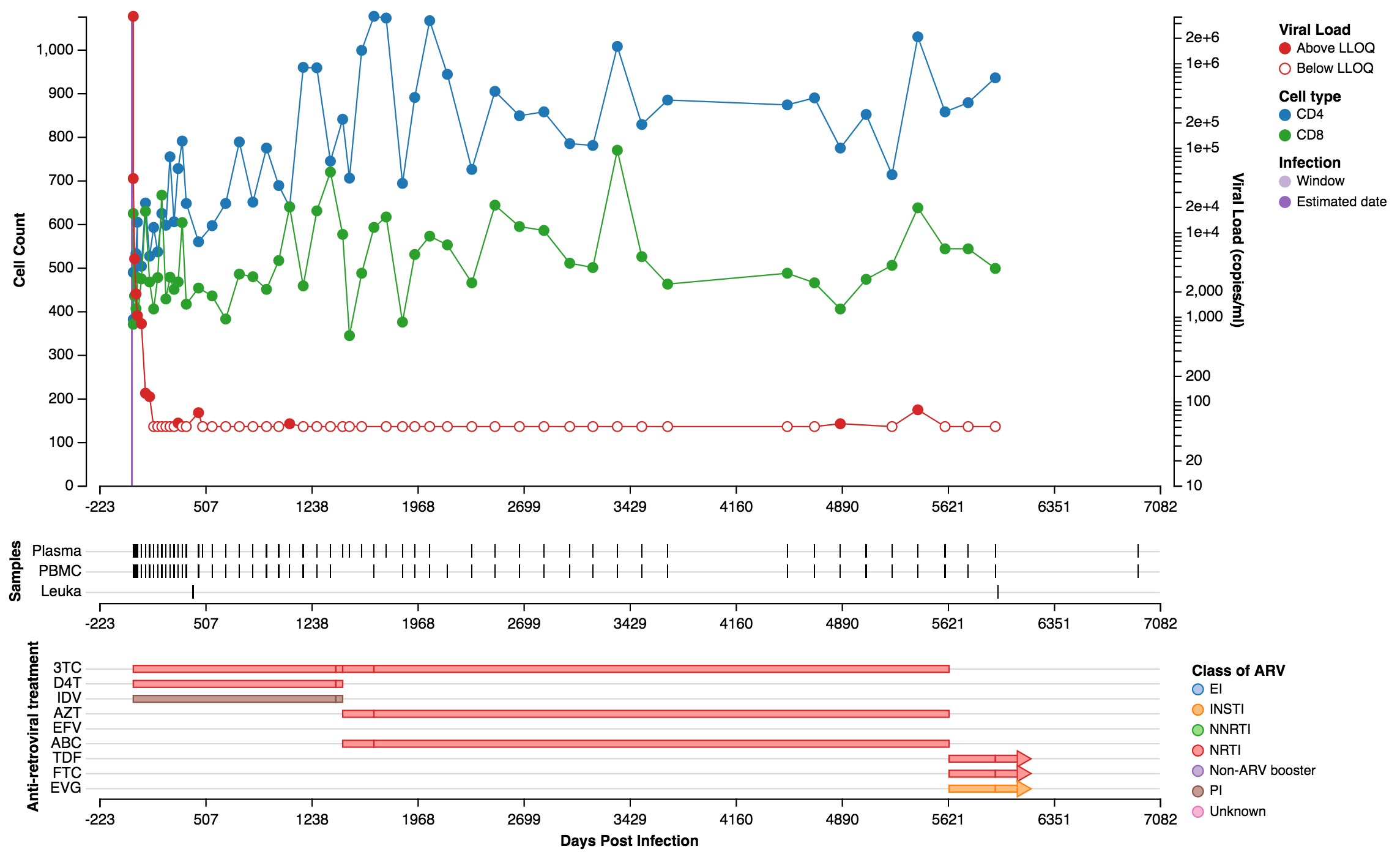
Import de-identified clinical data for research subjects, whether seen just once or continually over decades. Viroverse correlates samples with clinic visits and allows tracking of viral loads, T cell counts, and treatment regimens across time.
Freezer management
Viroverse builds in freezer management tools. Track tissue sample aliquots directly in Viroverse and easily look up box locations for retrieval. Know which specimens are available when planning new research.
History
Viroverse is developed in the laboratory of James Mullins. Early stages of development benefited from the involvement of Ram Samudrala and Roger Bumgarner and their laboratories.